Chapter 14 PCSF Module
The PCSF package performs fast and user-friendly network analysis using Prize-collecting Steiner Forest (PCSF) algorithm of high-throughput data by mapping the data onto a biological networks such as gene-gene, protein-protein interaction or any other correlation or coexpression based networks. Using the existing gene set or pathway database as a template, it determines high-confidence subnetworks relevant to the data, which potentially leads to predictions of enriched gene sets or pathways.
This module has 2 functional tabs: PCSF network and PCSF Function Cluster.
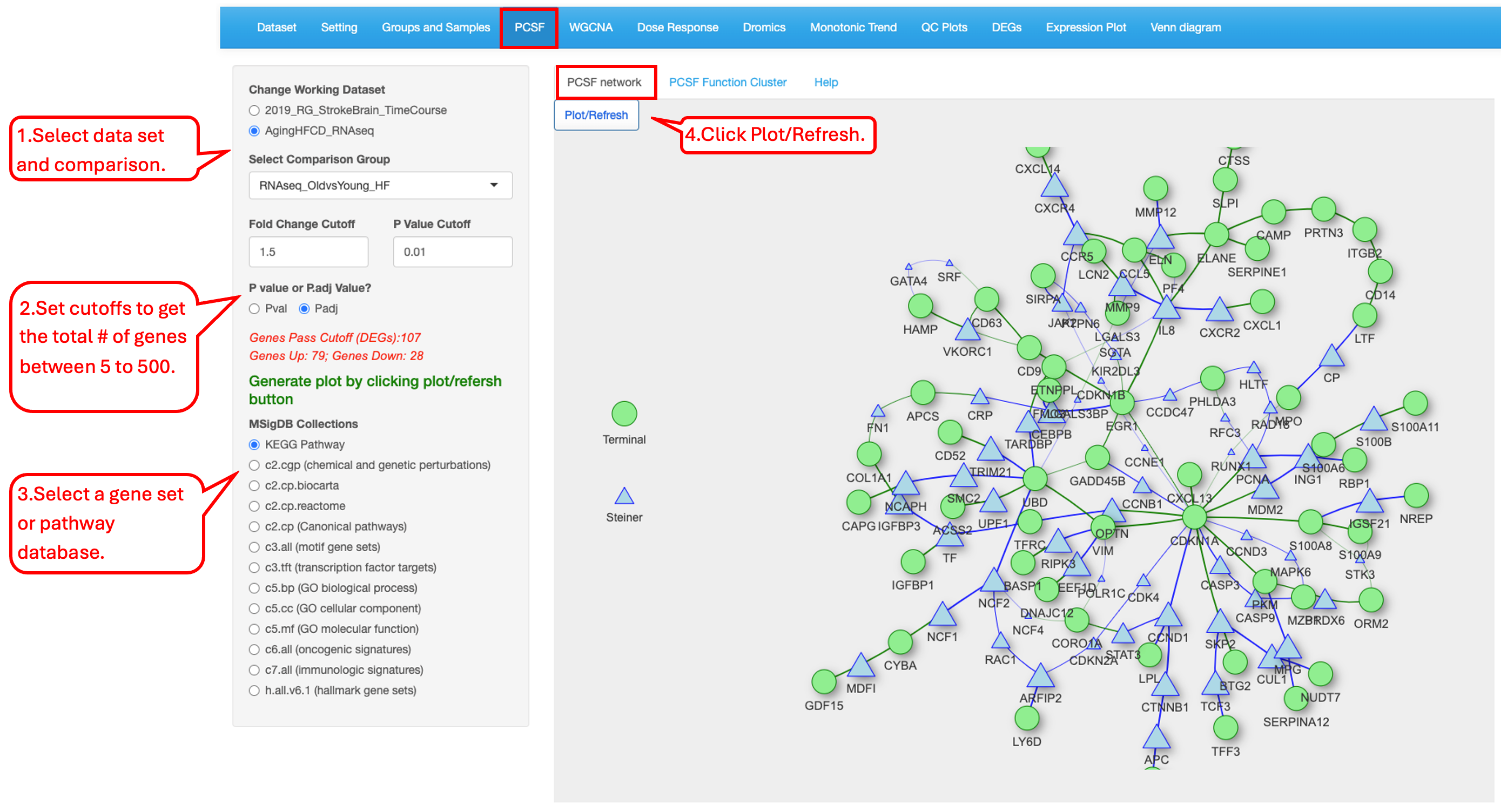
14.1 PCSF network
This tab shows the results of the computed high-confidence subnetwork relevant to the input data.
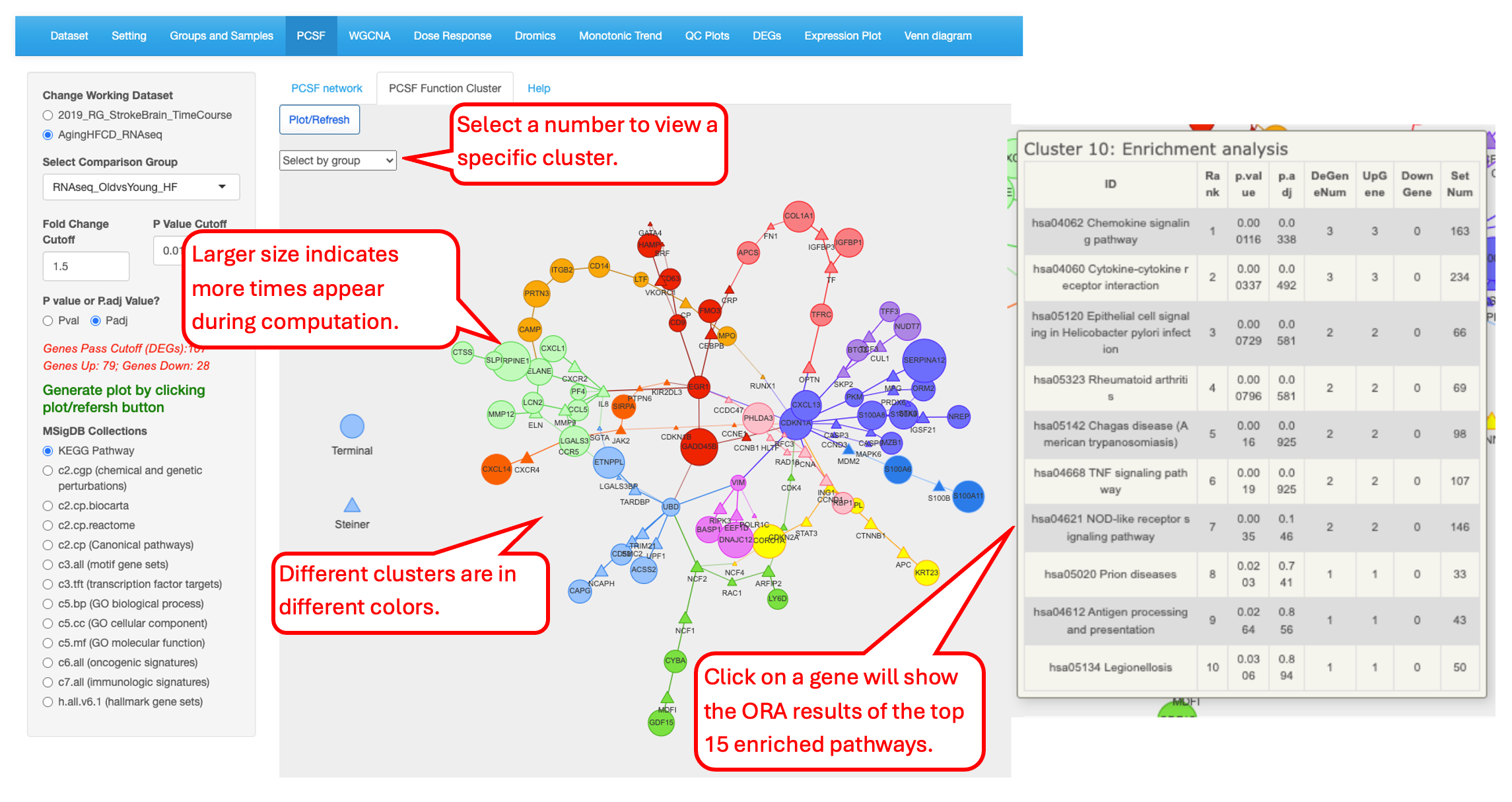
14.2 PCSF Function Cluster
This tab shows the interactive version of the annotated subnetwork. The node sizes and edge widths are proportional to the amount of times that node or edge appeared in the computation process. Nodes are colored by the clusters. Users can use mouse to click on a node in a cluster to view the table listing the top 15 functional enrichment pathways for the cluster which was calculated by the over-representation analysis(ORA). Each cluster can be visualized separately by “Select by group” icon located at the upper-left corner of the figure.