Chapter 9 Pattern Clustering Module
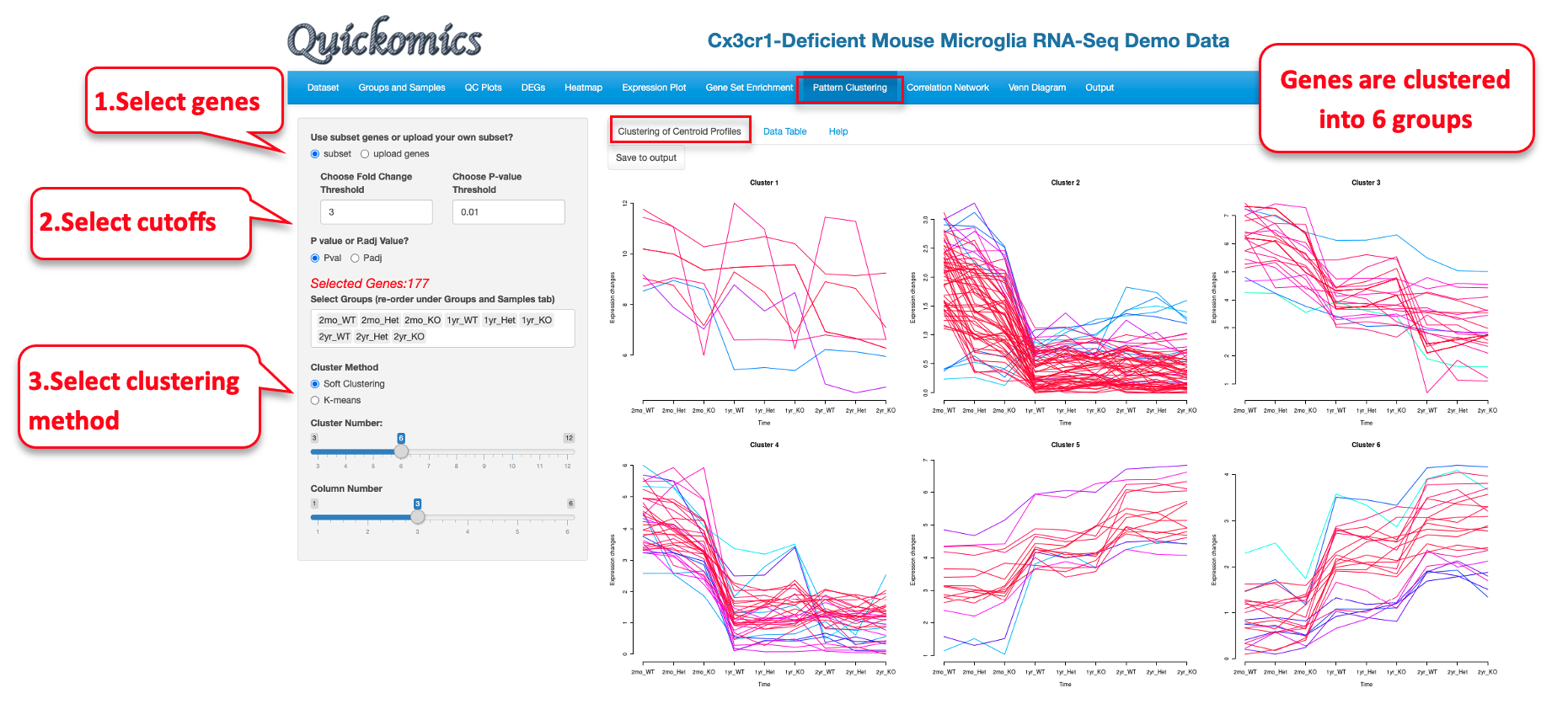
This functional module in Quickomics helps cluster genes/proteins based on their expression profiles across different groups. Three different algorithms, soft (fuzzy) clustering, k-means and partitioning around medoids (PAM), are implemented.
9.1 Clustering of Centroid Profiles
Users have the option to either select the DEGs/DEPS as the list of genes/proteins to cluster based off on or upload a custom gene list. DEGs/DEPs are selected based off all the comparisons, and cutoffs can be applied to limit the number of genes. In this example below, we use the top DEGs/DEPs with a Fold Change cutoff of 3 to identify 6 clusters of genes.
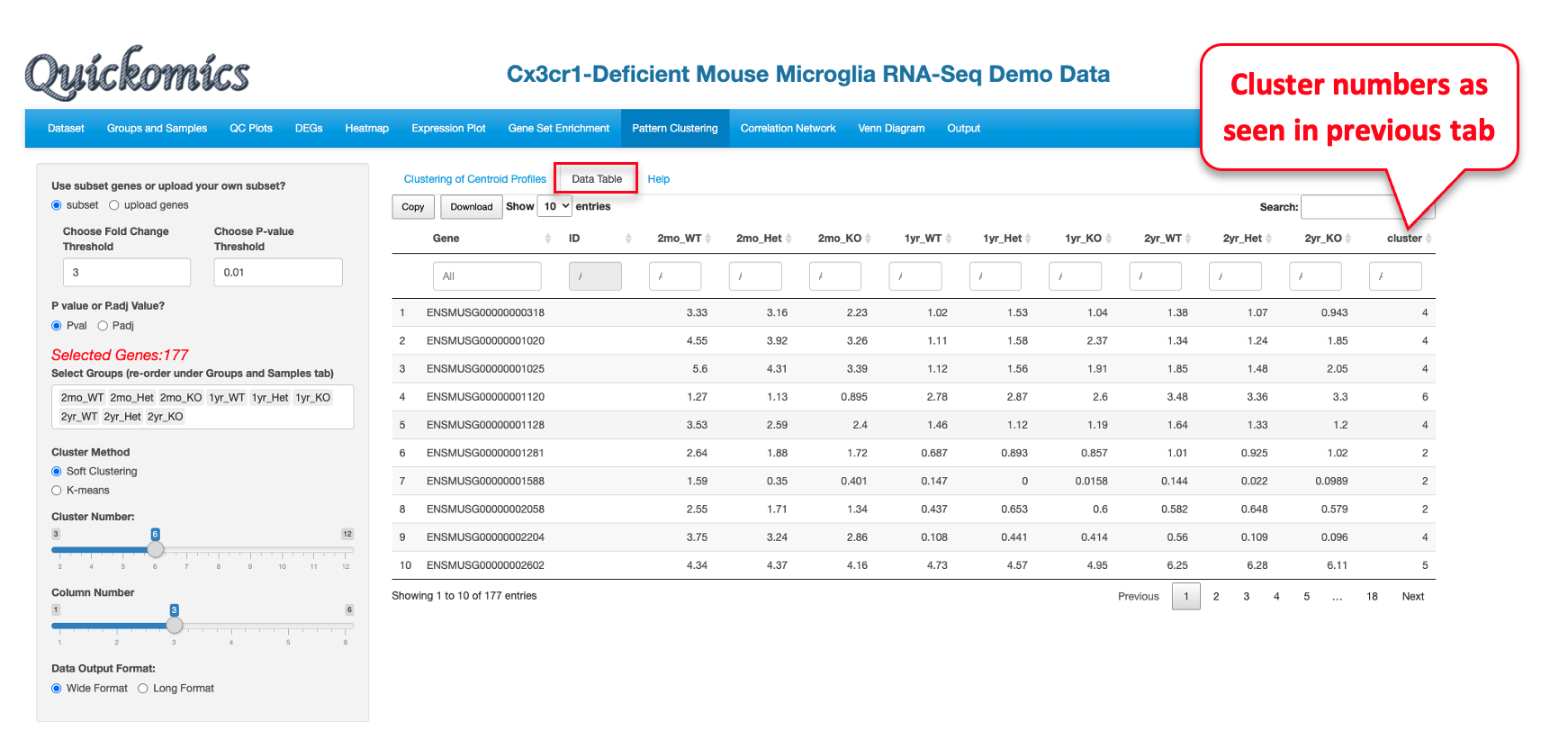
Please note that this visualization for this dataset is novel and was not previously reported in Gyoneva et al., 2019. The genes in each cluster can be viewed in the “Data Table” tab.