Chapter 4 Visium
10x Genomics Visium is a revolutionary technology that detects RNA expression and spatial location at the same time. Unlike traditional single-cell RNA-seq, which loses spatial information, Visium enables researches to map where gene activity occurs within the architecture of the intact tissue.
In detail, tissue sections can be placed on specialized Visium slides that contain an array of spatially barcoded capture spots. Each spot captures mRNA transcripts from the overlying tissue area, and at the same time, records the spatial location using the barcodes. After high-throughput sequencing, these data can be mapped back onto high-resolution tissue images, allowing researchers to analyze the gene expression patterns from distinct histological regions.
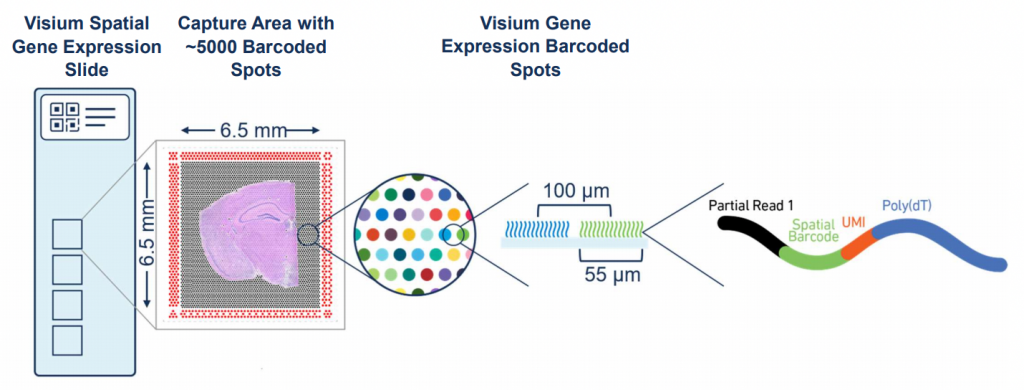
10x Genomics Visium slides and barcoded spots: As illustrated in the figure above, each 10x Genomics Visium slide has multiple captures (4 for regular 6.5x6.5 mm, or 2 for 11x11 mm). The barcoded spots are 55 μm in diameter, with gaps between each spot. (Image from: https://www.10xgenomics.com/)
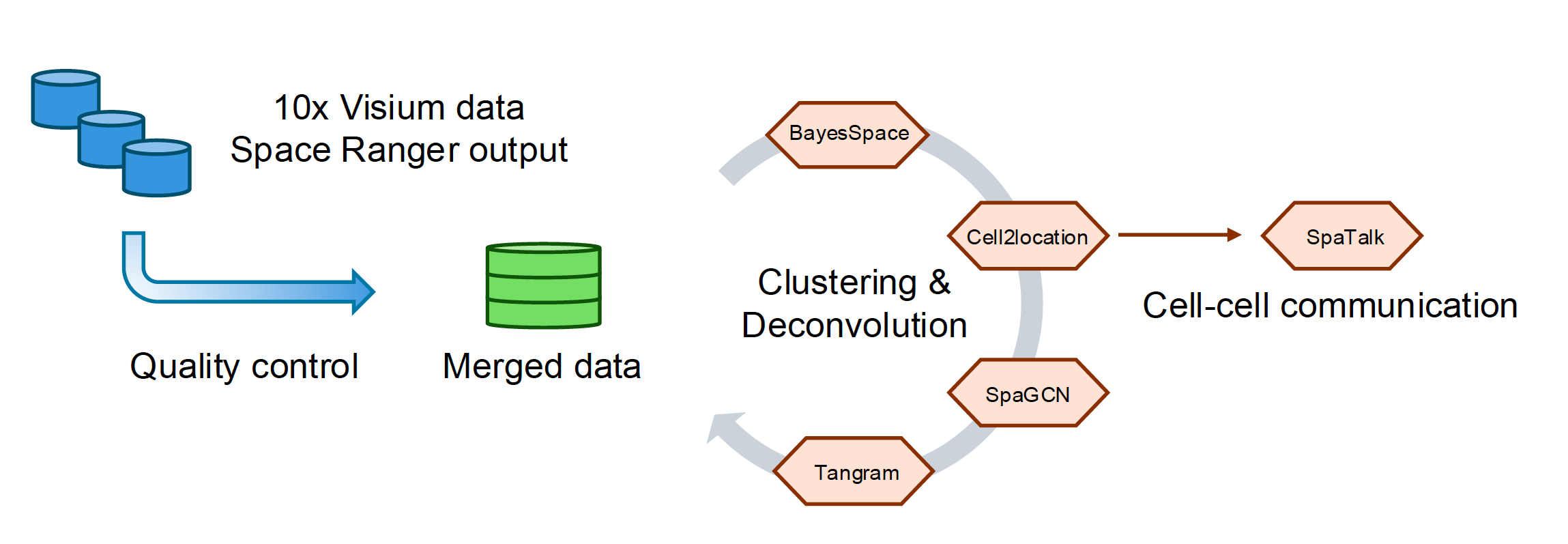
SpaceSequest Visium workflow: The Visium pipeline takes Space Ranger outputs and performs a series of analysis using multiple state-of-the-art tools, including BayesSpace, Cell2location, SpaGCN, Tangram, and SpaTalk.
4.1 Space Ranger run
If you start the analysis from FASTQ raw data, Space Ranger is necessary to process the data to gene expression count matrix, with spatial barcodes resolved. As we mentioned before, Space Ranger requires FASTQ files, high-resolution images, as well as key files (genome index, etc.) to run. We have illustrated these steps using a publicly available Visium data from an Alzheimer’s disease (AD) mouse model paper, with GEO entry: GSE203424. See the data preparation section 3.1.
A typical Space Ranger run command for Visium:
spaceranger count --id="WT-CO1" \
--transcriptome=~/Data/Reference_files/refdata-gex-mm10-2020-A \
--fastqs=~/Data/Fastq_files \
--cytaimage=~/Data/Sample1.tif \
--probe-set=~/Data/Reference_files/Visium_Mouse_Transcriptome_Probe_Set_v2.0_mm10-2020-A.csv \
--slide=H1-ABDCEFJ \ #Using a random slide ID for illustration
--area=A1 \
--localcores=4 \
--localmem=256 \
--create-bam=falseThis will generate the Space Ranger results that can be used to run SpaceSequest:
~/SpaceSequest_demo/1_Visium/
├── Input_data/
└── Demo_PSAPP.3Rep.Azimuth.Reference.h5ad #Downloaded from Azimuth website
└── SpaceRanger/
├── PSAPP-CO1/
├── ...
└── out/
├── PSAPP-CO1/
├── PSAPP-TAM1/
└── PSAPP-TAM1/4.2 Pipeline run
Here, we use the following run directory as an example: ~/SpaceSequest_demo/1_Visium
Demo data: 10x Visium data are from mouse brain (paper, and GEO record). Also, we downloaded the Azimuth mouse brain cortex reference for cell type deconvolution, with the following link: https://seurat.nygenome.org/azimuth/demo_datasets/allen_mop_2020.rds.
The first step is to generate the config and sampleMeta files by running the visium command by passing the working directory.
visium ~/SpaceSequest_demo/1_VisiumAfter this step, users can fill in the config.yml file and the sample.csv file. Please pay attention to the required items in the config.yml file as they are essential for the pipeline to run.
#Highlighting several key items:
prj_name: Visium_demo #Prefix of the final output
output: ~/SpaceSequest_demo/1_Visium/Run_pipeline #Output directoy will be created by the pipeline
methods: [SpaGCN,BayesSpace,cell2location,tangram,SpaTalk] #Software to run
#cell2location setting. This is for cell type deconvolution using a reference single-cell RNA-seq data
scH5ad: ~/SpaceSequest_demo/1_Visium/Input_data/Demo_PSAPP.3Rep.Azimuth.Reference.h5ad
annotation_obs: Cell_type
# tangram setting. This is for cell type deconvolution to map scRNA-seq to the spatial data and image
tg_scH5ad: ~/SpaceSequest_demo/1_Visium/Input_data/Demo_PSAPP.3Rep.Azimuth.Reference.h5ad
tg_annotation_obs: Cell_type
# SpaTalk seting. This is for cell-cell communication analysis
st_scH5ad: ~/SpaceSequest_demo/1_Visium/Input_data/Demo_PSAPP.3Rep.Azimuth.Reference.h5ad
st_annotation_obs: Cell_type
#Parallel and memory setting
parallel: "slurm" #Use "slurm" for Edge HPC
core: 10
memory: 505G #if provided (e.g. 100G), it will be used to request cluster scheduler
jobID: j47 # please make sure this job id is different with your other projects which would be run at the same time
For the sample.csv file, the first two columns (Sample_Name,SpaceRanger_path) are required, and here we added two more columns:
Sample_Name,SpaceRanger_path,Treatment,Image_hires
PSAPP-CO1,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-CO1/outs,Control,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-CO1-image.png
PSAPP-CO2,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-CO2/outs,Control,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-CO2-image.png
PSAPP-TAM1,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-TAM1/outs,Treated,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-TAM1-image.png
PSAPP-TAM2,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-TAM2/outs,Treated,~/SpaceSequest_demo/1_Visium/SpaceRanger/PSAPP-TAM2-image.pngThen, move the config.yml and sample.csv into a new folder called Run_pipeline, and run the visium script by passing the config.yml parameter to it:
#Run the pipeline
visium ~/SpaceSequest_demo/1_Visium/Run_pipeline/config.ymlAmong the software used in the pipeline, SpaTalk may need more memory and time. By testing the above four datasets, 500 Gb was sufficient for SpaTalk in a test run, but 250 Gb resulted in an Out-of-memory (OOM) error.
4.3 Results
Finally, users can check the results in the output directory. For each method, SpaceSequest generates a folder to store the results, for example: BayesSpace, SpaGCN, SpaTalk, etc. If there were any errors occurred, users can check the jxx (in this example, j47) folder to see the original scripts and error messages.
Output directory:
~/SpaceSequest_demo/1_Visium
├── BayesSpace/
├── ...
├── Visium_demo.pdf
├── Visium_demo_raw.rds
└── Visium_demo.rds
├── c2l/
├── model/
├── full_sc_model.h5ad
└── full_sc_model.pdf
├── PSAPP-CO1.h5ad
├── PSAPP-CO1.pdf
├── PSAPP-CO1.pkl
├── PSAPP-CO2.h5ad
└── ...
├── SpaGCN/
├── align.pdf
├── align.pkl
├── SpaGCN.pdf
└── ...
├── SpaTalk/
├── PSAPP-CO1/ #Multiple figures in each folder associated with each sample name.
├── PSAPP-CO2/
├── PSAPP-TAM1/
├── PSAPP-TAM1/
└── Visium_demo_C2L.h5ad
├── logNormal/
└── Visium_demo.h5ad
├── tangram/
├── tmp
├── Visium_demo.pkl
├── Visium_demo_sc_location.h5ad
└── Visium_demo_segment.h5ad
├── QC/
├── metrics_summary.csv
└── QC.pdf
├── config.yml
├── config.yml.20250719.log
└── sample.csvExample outputs:
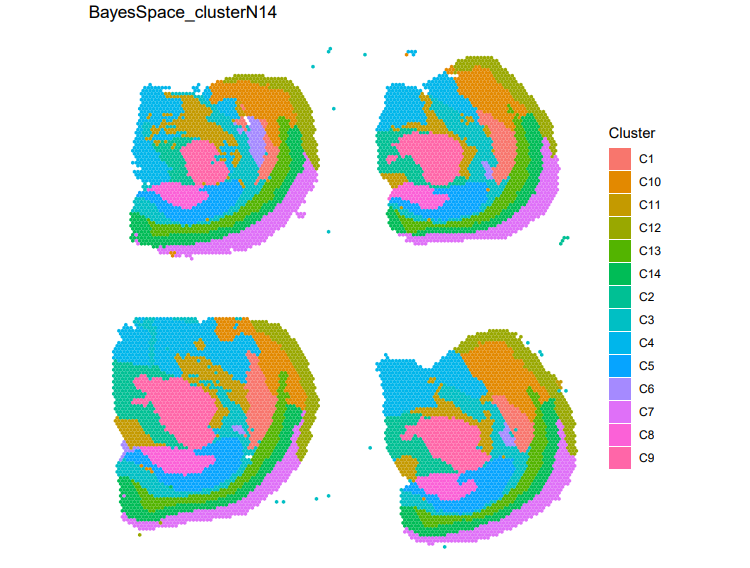 Clustering results generated by BayesSpace for the Visium spatial transcriptomics dataset.
Clustering results generated by BayesSpace for the Visium spatial transcriptomics dataset.
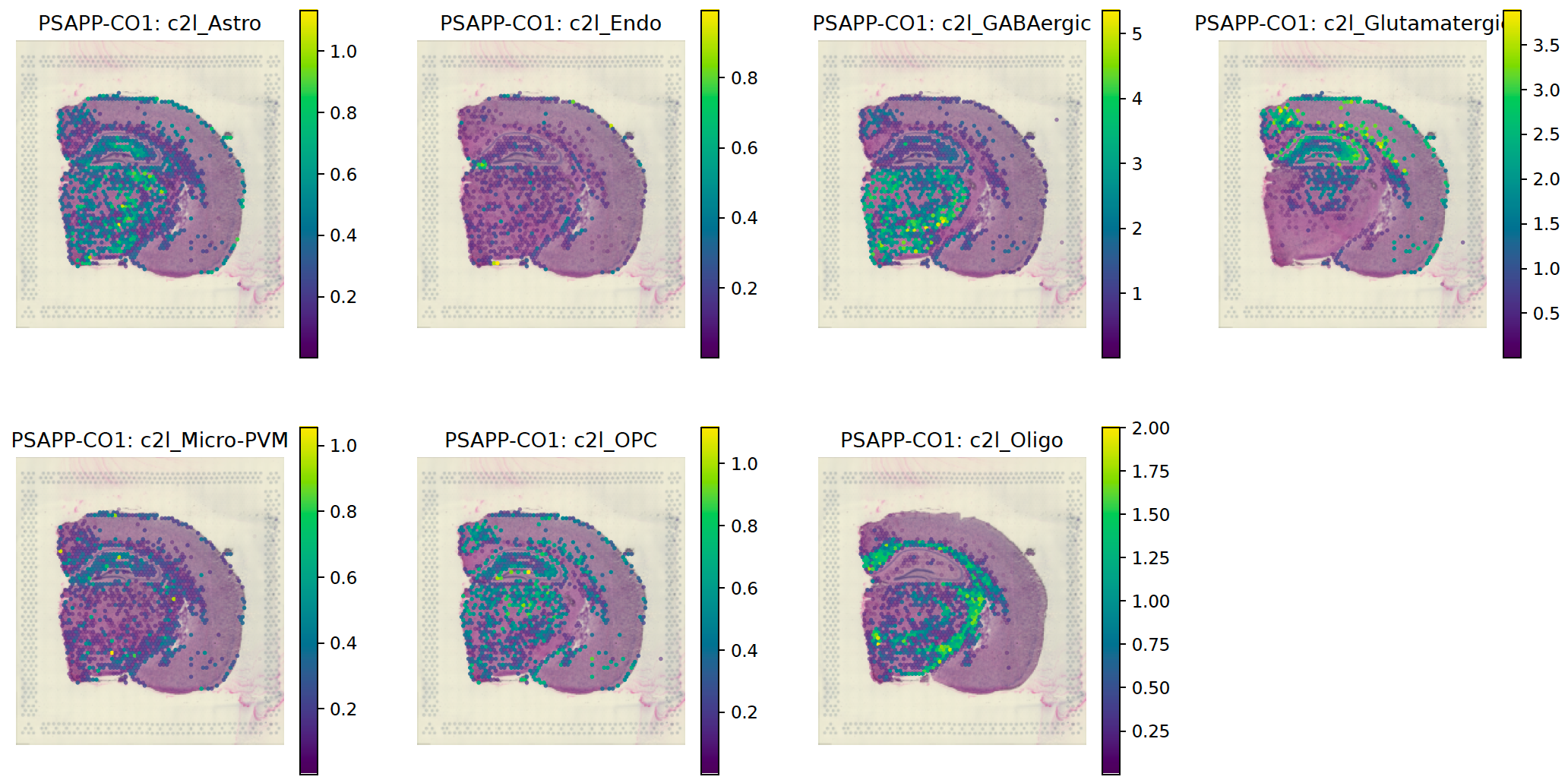 Cell2location result for cell type deconvolution using single-cell RNA-seq data, guided by a reference downloaded from Azimuth.
Cell2location result for cell type deconvolution using single-cell RNA-seq data, guided by a reference downloaded from Azimuth.