Chapter 11 ShinyOne component
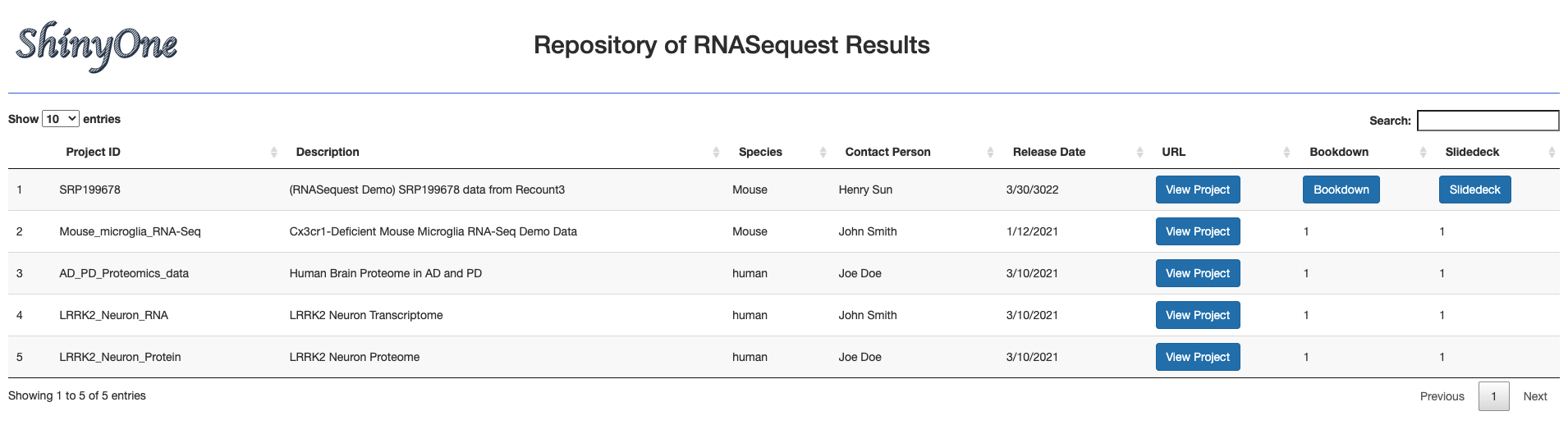
We also developed ShinyOne, which is an R Shiny App displaying a web-based searchable table that lists all projects processed by RNASequest and loaded into Quickomics, with Quickomics launching link for each project.
In the backend of Quickomics, we added the capability to load projects directly from URLs after the data files have been saved at user-specified server locations. Together, ShinyOne and URL loading enable many projects to be powered by one Quickomics instance.
Please use the following link to access ShinyOne:
http://shinyone.bxgenomics.com
To add a new project to ShineOne, one can simply add one line to the project.csv file in the ShinyOne App folder. For example, after processing project SRP199678 using the EA pipeline, we have the following:
QuickOmics Visualization Link:
SlideDeck:
https://interactivereport.github.io/RNASequest/ShinyOne/SRP199678/SlideDeck
Bookdown report:
https://interactivereport.github.io/RNASequest/ShinyOne/SRP199678/Bookdown
Here is an example of adding this information to the project.csv file:
| Project ID | Description | Species | Contact Person | Release Date | URL | Bookdown | Slidedeck | |
|---|---|---|---|---|---|---|---|---|
| SRP199678 | SRP199678 | (RNASequest Demo) SRP199678 data from Recount3 | Mouse | Henry Sun | 3/30/2022 | https://quickomics.bxgenomics.com/?serverfile=SRP199678 | https://interactivereport.github.io/RNASequest/ShinyOne/SRP199678/Bookdown | https://interactivereport.github.io/RNASequest/ShinyOne/SRP199678/SlideDeck |
And it will show up in the ShinyOne Page: