Chapter 7 Advanced plotting functions
For advanced users, EA offers versatile plotting functions to adjust your figures. Each plotting function within Plot_Functions.R has many options, mirroring the user interface from QuickOmics. Here are several examples:
7.1 PCA plot
# Load RData created from EArun
# Here is an example
# https://github.com/interactivereport/Quickomics/blob/master/data/Mouse_microglia_RNA-Seq.RData
load("data/Mouse_microglia_RNA-Seq.RData")
# Load Plot_Functions.R
# https://github.com/interactivereport/RNASequest/tree/main/src
setwd("~/RNASequest/src/")
source("Plot_Functions.R")
# Create a basic PCA Plot.
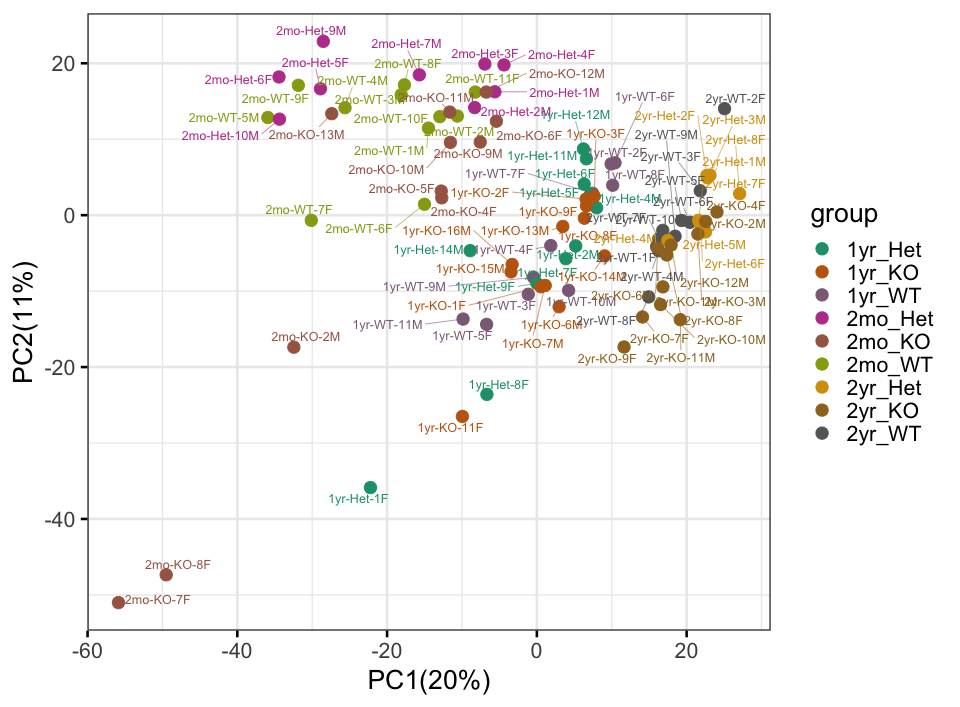
p<-PCA_Plot(data_wide, MetaData)
print(p) # PCA plot with additional options:
p<-PCA_Plot(data_wide, MetaData, pcnum=c(2, 3),label_samples=F,
PCAcolorby="Age", PCAshapeby="Genotype", ellipsoid=T, rug = T)
# Plot only selected samples:
input_samples=MetaData$sampleid[20:60]
p<-PCA_Plot(data_wide, MetaData, input_samples)
print(p) 7.2 Covariate checking
# Check covariates:
# This can be displayed as a table in R Markdown
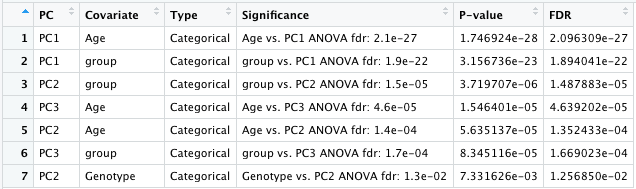
res<-PC_covariates_out(data_wide, MetaData)
sig_covariates=res$selVar_All #data frame to print as table# Check covariates with more options if needed:
res<-PC_covariates_out(data_wide, MetaData, input_samples, select_covariates=c("Age", "Genotype", "Gender"), PC_cutoff=3, FDR_cutoff=0.2)
(sig_covariates=res$selVar_All) 7.3 Volcano plot
# Loop through the comparisons.
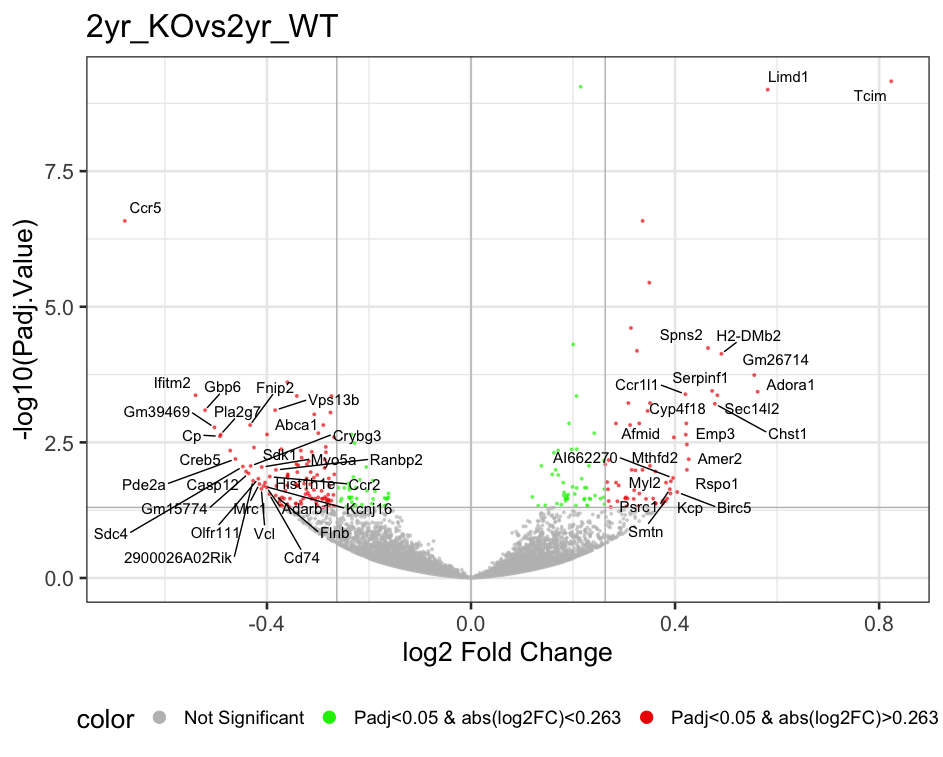
p<-Volcano_Plot(results_long, ProteinGeneName, comparison="2yr_KOvs2yr_WT")
print(p) # More controls on volcano plot:
p<-Volcano_Plot(results_long, ProteinGeneName, comparison="1yr_KOvs1yr_WT", Ngenes=40, rasterize="No", vlegendpos="right", Max_Pvalue=30, Max_logFC=1.5) 7.4 Heatmap
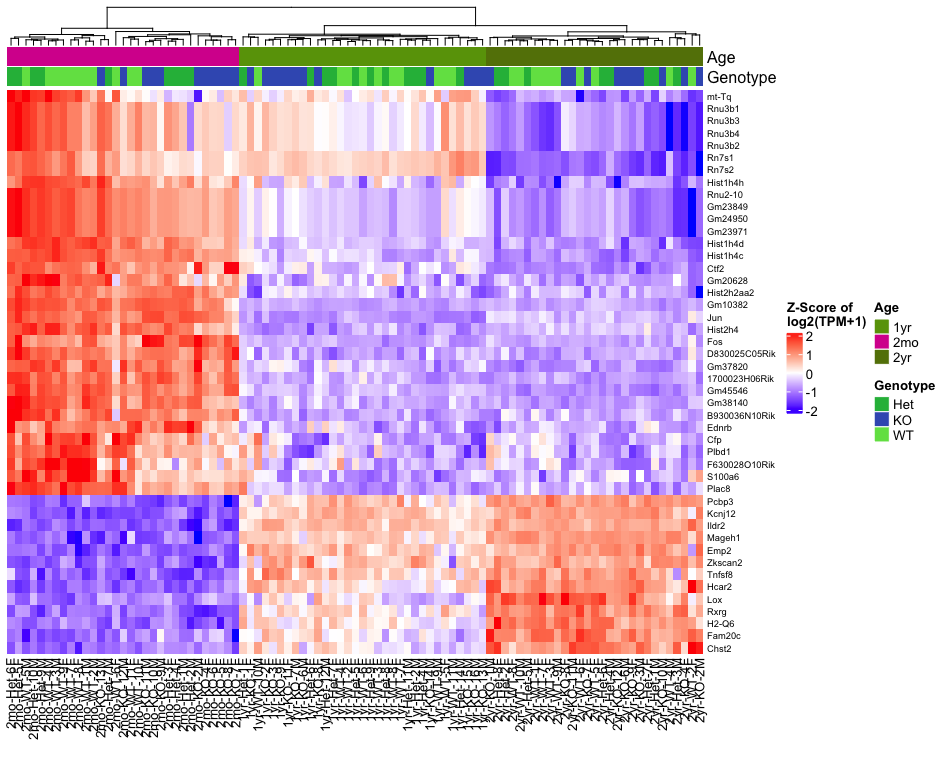
# Heatmap
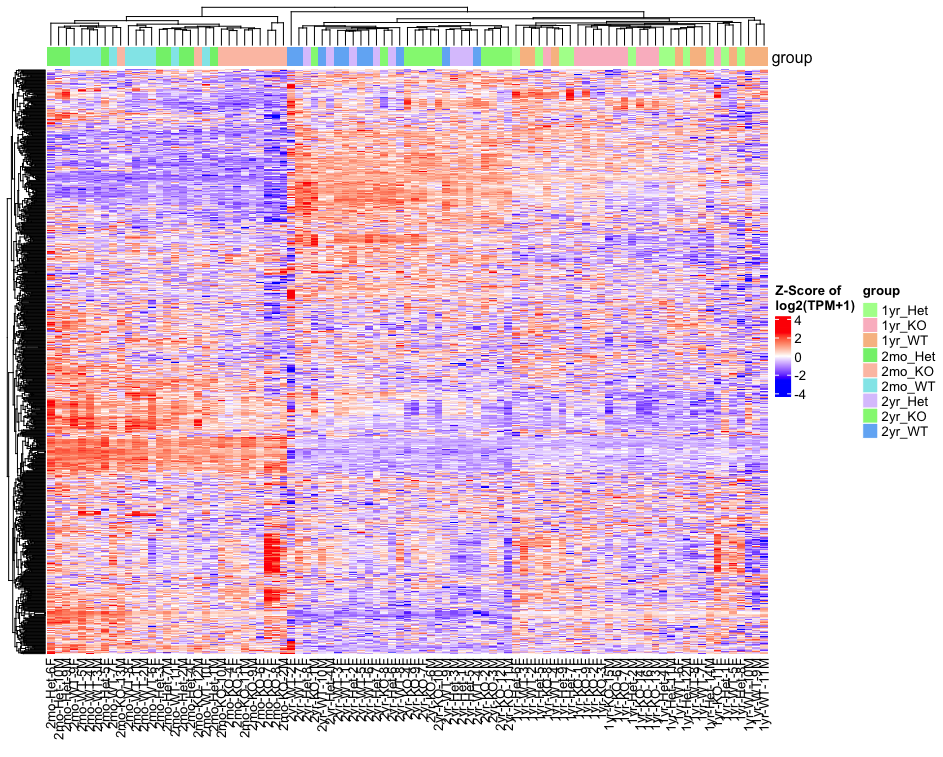
# Overview from 1000 most variable genes
p<-Heatmap_Plot(data_wide, MetaData, results_long, ProteinGeneName,heatmap_subset="variable", maxgenes=1000)
draw(p, merge_legend=T, auto_adjust = FALSE) # Heatmap from DEGs of a comparison:
# This can be done for all comparisons.
p<-Heatmap_Plot(data_wide, MetaData, results_long, ProteinGeneName,heatmap_subset="subset", comparison="2yr_WTvs2mo_WT")
draw(p, merge_legend=T, auto_adjust = FALSE) # With more options to customize the heatmap
p<-Heatmap_Plot(data_wide, MetaData, results_long, ProteinGeneName,heatmap_subset="subset", comparison="2yr_WTvs2mo_WT", heatmap_fccut=4, heatmap_pvalcut=0.01, heatmap_psel = "Padj", heatmap_row_dend=FALSE, heatmap_annot=c("Age", "Genotype") )
draw(p, merge_legend=T, auto_adjust = FALSE)